We are planning several exciting workshops for SB3C 2024. Please check back often as details will be updated as they become available.
Tuesday, June 11th from 3:00 – 4:00pm:
Lance Frazer, Southwest Research Institute
Manoj Myneni, W.L. Gore & Associates
Unsure of what a job outside of academia looks like? Unsure of how you could fit into the workforce with an advanced degree? It may seem difficult to navigate the transition between academia and industry, but it doesn’t have to be. With a little bit of information and perspective from those that have gone on to industry after graduating with an advanced degree, you’ll be ready and encouraged to look for opportunities beyond graduation. This workshop is designed to give students an insight into industry jobs. What skills from academia are important, what lessons we’ve learned, what we wish we would’ve known, and what you can start doing today to better prepare yourself for the job search. The session will include two brief talks followed by a Q&A panel of several industry leaders. Come join us and learn what the next steps of your career could look like!
Wednesday, June 12th from 3:45 – 5:15pm:
Spencer Szczesny, Pennsylvania State University
Darryl Dickerson, Florida International University
It is well established that all individuals harbor implicit biases/prejudices that influence our perspectives, opinions, and decisions. In many cases, these biases are innocuous heuristics that are necessary to navigate the numerous choices we make on a daily basis (e.g., choosing a name-brand medication over a generic). However, often these biases influence larger decisions and behaviors that negatively impact others and reinforce existing social inequalities. A particularly relevant example in science is the peer review process, which is the standard for determining what research gets funded and which papers get published. While many reviewers and editors may attempt to be as objective as possible, there is clear evidence that peer review is an inherently subjective determination influenced by many forms of bias. Therefore, the goal of this workshop organized by the ASME Journal of Biomechanical Engineering (JBME) is to broaden awareness of bias at all levels within the peer review process and inform attendees of the best practices for mitigating its impact on publication decisions.
Participants will learn about efforts being implemented within JBME to minimize bias in paper selection as well as hear from outside experts about their research on the various forms of bias in peer review and evidence-based mitigation strategies. This workshop will help dismantle the invisible obstacles created by implicit biases in identifying the best ideas to fund and the best science to disseminate. Additionally, it will help create a scientific community that is more educated about the effects of bias on the peer review process and the best steps to mitigate its negative impact. Finally, attendees will be better equipped to identify whether a journal properly addresses bias in the peer review process so that their work is fairly assessed.
Thursday, June 13th from 2:00 – 3:30pm:
 Corinne Henak, University of Wisconsin-Madison
Corinne Henak, University of Wisconsin-Madison
Sara Wilson, University of Kansas
Katie Knaus, Colorado School of Mines
Jacob Merson, Rensselaer Polytechnic Institute
The goal of this workshop is to provide participants with knowledge, hands-on experience, and discussion about using generative artificial intelligence (AI) in education and research. This workshop builds from a series of virtual ASME BED workshops held in fall 2023, which had the topics of Overview and Ethics; Applications in Education; and Applications in Research. The workshop will begin with a recap of themes brought up in the virtual workshops, along with updates in the rapidly changing landscape from fall 2023 to summer 2024. The majority of the workshop will be spent in hands-on exploration of the power and limitations of generative AI, using the tools to work on topics including: teaching or debugging code for introductory classes; writing a syllabus; writing a portion of a research paper; and writing a teaching statement for a faculty application. At the end of the session, groups will write white-papers and debrief on their findings.
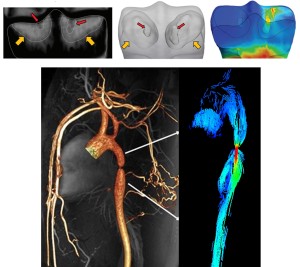 Alejandro Roldán-Alzate, University of Wisconsin-Madison
Alejandro Roldán-Alzate, University of Wisconsin-Madison
Corinne Henak, University of Wisconsin-Madison
Colleen Witzenburg, University of Wisconsin-Madison
Josh Roth, University of Wisconsin-Madison
Stephanie Cone, University of Delaware
Solution to Biomechanics, Bioengineering, & Biotransport problems require a multidisciplinary approach. A very important aspect is the actual “user” of the solutions proposed by the engineering team which are the clinical doctors. In this workshop we will hear from clinicians from different areas including fluid dynamics and solid mechanics who will tell us about ways in which engineering has been useful for them in the clinic.
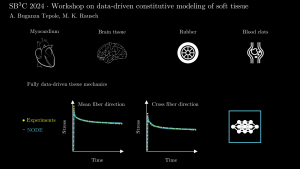 Adrian Buganza Tepole, Purdue University
Adrian Buganza Tepole, Purdue University
Manuel K Rausch, The University of Texas at Austin
Machine learning (ML) has permeated into all areas of engineering and tissue modeling is no exception. However, alongside the democratization of ML tools through open source packages (e.g., Pytorch) and cloud computing (e.g., Google Colab), there is a need for educational materials, including demos and benchmarks. This workshop fills that need. The goal of this workshop is to give an overview of data-driven methods to model soft tissues that might be of interest to the biomechanics community attending SB3C, and to give hands-on examples using Python Jupyter notebooks on the Google Colab with synthetic and experimental data. The workshop will start with an overview of current methods in the literature as well as the type of experimental data needed to train data-driven methods. Then, hands-on examples will show how to train the models on the available data and how to evaluate the model for stress prediction for a given deformation. Application into finite element software will also be discussed. The methods that will be presented can be used for a variety of materials (skin, rubber, blood clots, myocardium, brain) and for a variety of phenomena (hyperelasticity, viscoelasticity, damage). Particular attention will be given to methods that guarantee physics constraints a priori, just as the most common models for soft tissues do, thus allowing for flexibility while retaining physical behavior.
Anita Singh, Temple University
This workshop will focus on raising awareness and informing the audience of FDA strategies that serve to promote and protect the health of diverse populations through research and communication of science that addresses healthcare disparities. Additional topics will include details of how to design a new medical device and get it approved for sale. Brief presentations on product design and development processes used in the development of medical devices, getting FDA approval for the device, and where to get help will also be offered. Additionally, the finalists of the NSF-funded UG Design Competition held at SB3C will have a hands-on component aimed at improving upon their design ideas while accounting for issues related to healthcare disparity such as expanding the stakeholder community, understanding diverse patient perspectives, preferences, and unmet needs, and how to design a killer experiment that accounts for enrollment of underrepresented populations etc.
Friday June 14th from 8:30 – 11:30 am
C. Alberto Figueroa, University of Michigan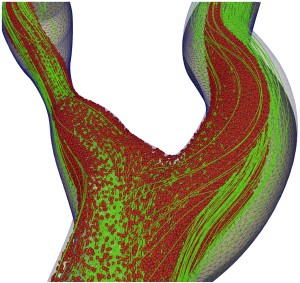
Abhilash Malipeddi, University of Michigan
Jesse Capecelatro, University of Michigan
In biofluids, the Digital Twin paradigm relies on leveraging medical images and pressure and flow data to perform customized, subject-specific predictions. Flows in blood vessels and lymphatic systems include complex structured fluids, composed of a liquid phase (plasma) and a disperse phase, which includes cells and other particles such as thrombus of vastly different sizes and biophysical properties, and interactions between particles, fluid, and vessel boundaries. The development of robust and scalable schemes to study these complex interactions is a challenging task.
We recently developed a volume-filtered Eulerian-Lagrangian strategy that uses a finite element method (FEM) to solve for the fluid phase coupled with a discrete element method (DEM) for the particle phase.
In this workshop, we will demonstrate this FEM-DEM formulation for performing efficient and scalable particle-laden flow simulations for digital twinning in biofluids, implemented in the open-source framework CRIMSON.
Jeffrey Weiss, University of Utah
Gerard Ateshian, Columbia University
In this workshop, participants will learn how to use FEBio and FEBio Studio from the people who have been developing the software since 2007. From its inception, the software has continued to grow with a focus on applications in biomechanics and biophysics, and we hope to give the audience an overview of the current capabilities, new developments, and future directions. We will also discuss the use of FEBio and FEBio Studio for teaching in biomedical engineering courses.
First, we will give an overview of the progress on several new development projects, such as import, visualization, and integration of 3D image data in a finite element model; FEBio plugin integration and generation in FEBio Studio; Python scripting with FEBio Studio, and more.
Then, we will lead a hands-on workshop. Participants will learn the basics of setting up and running models in FEBio Studio. They will learn how to import and create geometry, apply materials, boundary conditions, and loads that define the physics of the problem. In addition, they will try out different contact algorithms and learn how to decide which one works best for a particular problem. Next, they will learn how to generate the meshes, and how to generate mesh-dependent scalar and vector fields that can be used in many of the model components, such as generating fiber distributions. Finally, participants will run the models on their own laptops and visualize the results. The workshop will be mostly geared towards beginner users, but the many new features in the code will ensure that even intermediate and advanced users will learn something new.
Finally, a Q/A session will wrap up the workshop and participants are encouraged to bring their models and discuss their challenges with the presenters.
Andrew Anderson, University of Utah
Bergen Braun, University of Utah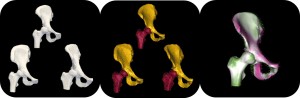
Researchers in biology, engineering, and medicine commonly use form (i.e., shape) to understand function. In these fields, it is understood that abnormal morphology of the underlying anatomy often leads to impaired function – this is certainly true of the musculoskeletal system. While analysis of tissue shape from medical imaging is central in diagnosis and treatment planning, clinical observations of shape are often qualitative since quantitative description of shape requires the application of mathematics, statistics, and computing to parse the shape into a numerical representation. ShapeWorks is an open-source software that quantifies population-level shape representation derived from 3D tissue reconstructions from imaging data. ShapeWorks is integrative, user-friendly, and scalable, and its utility has been demonstrated across a range of biomedical engineering applications. This workshop aims to introduce ShapeWorks to the SB3C Community through a description of the core algorithms and a presentation of published shape models of human tissues. More advanced functionality, including multidomain modeling and statistical parametric mapping of features that accompany shape will be discussed by live demo. Ongoing research and development efforts will also be introduced, including measures we are taking to increase efficiency and broaden the application of shape analyses through the use of machine learning.
Alison Marsden, Stanford University![]()
David Parker, Stanford University
Shawn Shadden, UC Berkeley
Vijay Vedula, Columbia University
Martin Pfaller, Yale University
Nathan Wilson, Open Source Medical Software Corporation
Description: SimVascular is a fully open-source software package providing a complete pipeline from medical image data to cardiovascular blood flow simulation results and analysis (www.simvascular.org). It offers capabilities for image segmentation, unstructured adaptive meshing, physiologic boundary conditions, and multi-physics simulations. The svFSIplus finite element solver incorporates fluid structure interaction capabilities, including large deformation motion with an Arbitrary Lagrangian Eulerian (ALE) formulation, electrophysiology, and cardiac mechanics solvers. The solver has recently been released with a new C++ codebase. An accompanying vascular model repository (VMR, www.vascularmodel.com) provides over 250 freely available clinical data sets with image data and simulation results from different parts of the vascular anatomy. The VMR supports research in machine learning, medical devices, and reduced order modeling. Extensive online documentation and video tutorials with clinical examples are provided online.
In this workshop, we will offer focused sessions tailored to new and experienced users. New users will be guided through step-by-step tutorials, covering basic steps of model construction, meshing, flow simulations, and best practices (and pitfalls to avoid) for high quality results. For experienced users, we will cover advanced topics such as cardiac mechanics and electrophysiology, reduced order modeling, interactive surgical planning, and automated scripting via the Python interface. Users will have the opportunity to discuss current challenges from their research with the SimVascular developers and thus participants are encouraged to bring their own models and questions to the workshop.
Robb Colbrunn – Cleveland Clinic
Stephanie Cone – University of Delaware
Josh Roth – University of Wisconsin
Lesley Arant – University of Wisconsin
Emma Coltoff – Wake Forest Baptist Health
Tara Nagle – Cleveland Clinic
Elizabeth Pace – Cleveland Clinic
Jeremy Loss – Cleveland Clinic
Callan Gillespie – Cleveland Clinic
Many in vitro joint biomechanics researchers, and their in vivo and in silico collaborators, attend the SB3C conference but only participate in sessions regarding their specific joint or clinical problem of interest. Best practices, novel methodologies, and unique analysis techniques are not necessarily joint or clinical question specific. Researchers using simVITRO systems have expressed a desire for a workshop to collaborate and discuss these technical challenges and solutions with the greater biomechanical engineering community.
At this workshop we aim to present an overview of robot-based orthopedic biomechanics research to newcomers in the field; explaining the What, Why and How of 6 degree of freedom robotic in vitro joint testing, and to present more advanced topics relevant to seasoned researchers. We also want to provide in vitro, in silico, and in vivo joint biomechanics researchers the ability to network and discuss technical challenges and solutions for collecting in vitro joint biomechanics data. The workshop will include presentations by researchers working on novel solutions in this field, hands-on experience through robotic demonstrations, and break-out sessions for learning how to get the most out of your biomechanical testing system.
